Long non-coding RNAs (lncRNAs) were commonly viewed as non-coding elements. However, they are increasingly recognised for their ability to be translated into proteins, thereby playing a significant role in various cellular processes and contributing to the pathogenesis of diseases. With developments in biotechnology and computational algorithms, a range of novel approaches are being applied to investigate the translation of lncRNAs.
Here, we report the development of the LncPepAtlas database
(http://www.cnitbiotool.net/LncPepAtlas/), which aims to represent an integrated annotation platform for the upstream regulation of lncRNAs and to act as a resource compiling evidence for the translation of lncRNA-encoded peptides across multiple species.
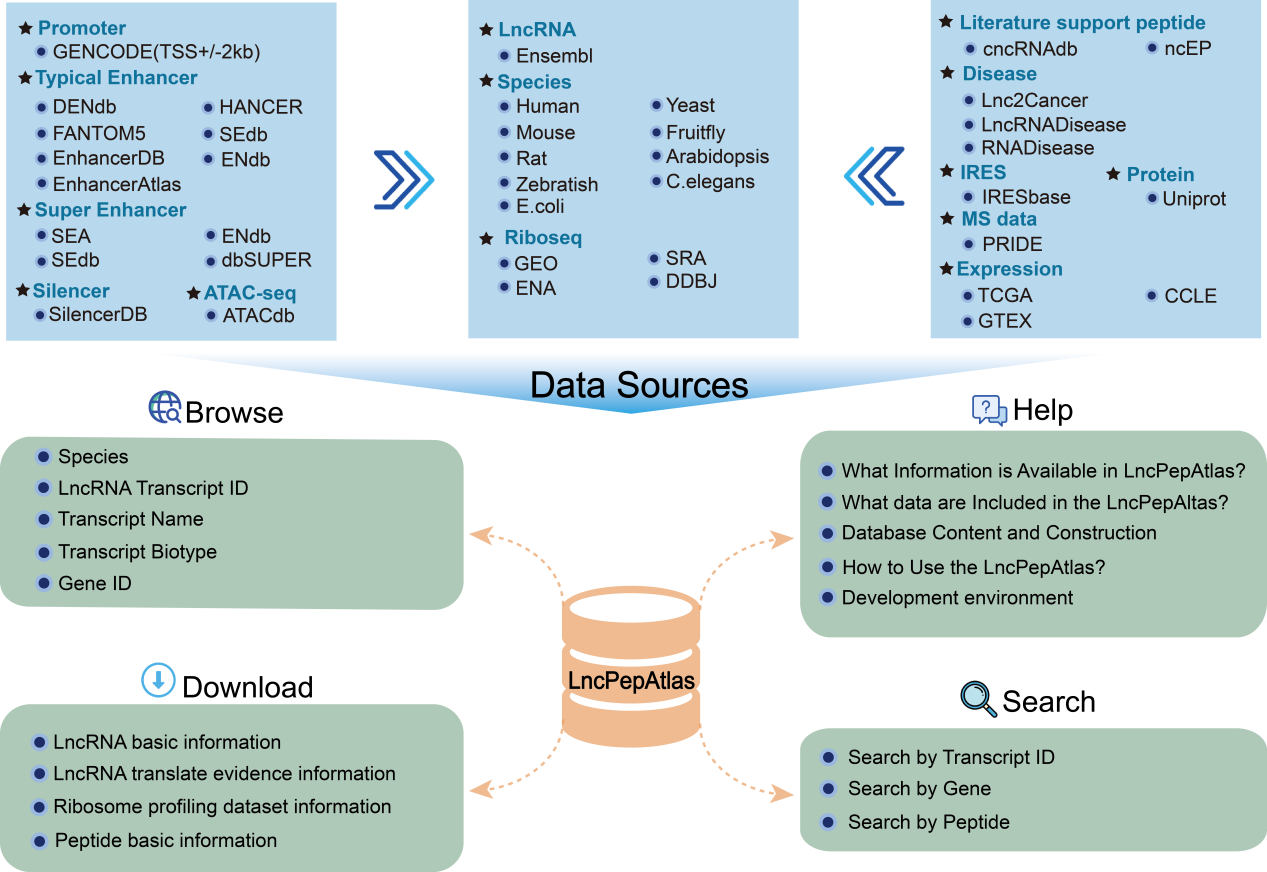
The platform compiles an extensive repository of translated lncRNA peptides, by analysing 2,655 ribosome profiling datasets from nine species, including human, mouse, rat, arabidopsis thaliana, caenorhabditis elegans, danio rerio, drosophila melanogaster, Escherichia coli and Saccharomyces cerevisiae. During the next step, nine methods were employed to assess the translational evidence of lncRNA-derived peptides from the Ensembl database. These included manual curation, ribosome occupancy, IRES, m6A, mass spectrometry, Pfam, CPC2, CPAT and CNCI.
In addition to exploring translational evidence, LncPepAtlas also offers extensive annotations for lncRNAs, that is intended to identify upstream regulatory elements such as transcription factors (TFs), super enhancers (SEs), enhancers and silencers across a diverse range of cancer types, tissues or cell lines. In addition,, LncPepAtlas also analyzes lncRNA-encoded peptides, including their alignment with experimentally validated protein sequences and providing an assessment of their binding affinities as antigenic peptides interacting with both Class I and Class II major histocompatibility complex (MHC) molecules.
4.1 Browse
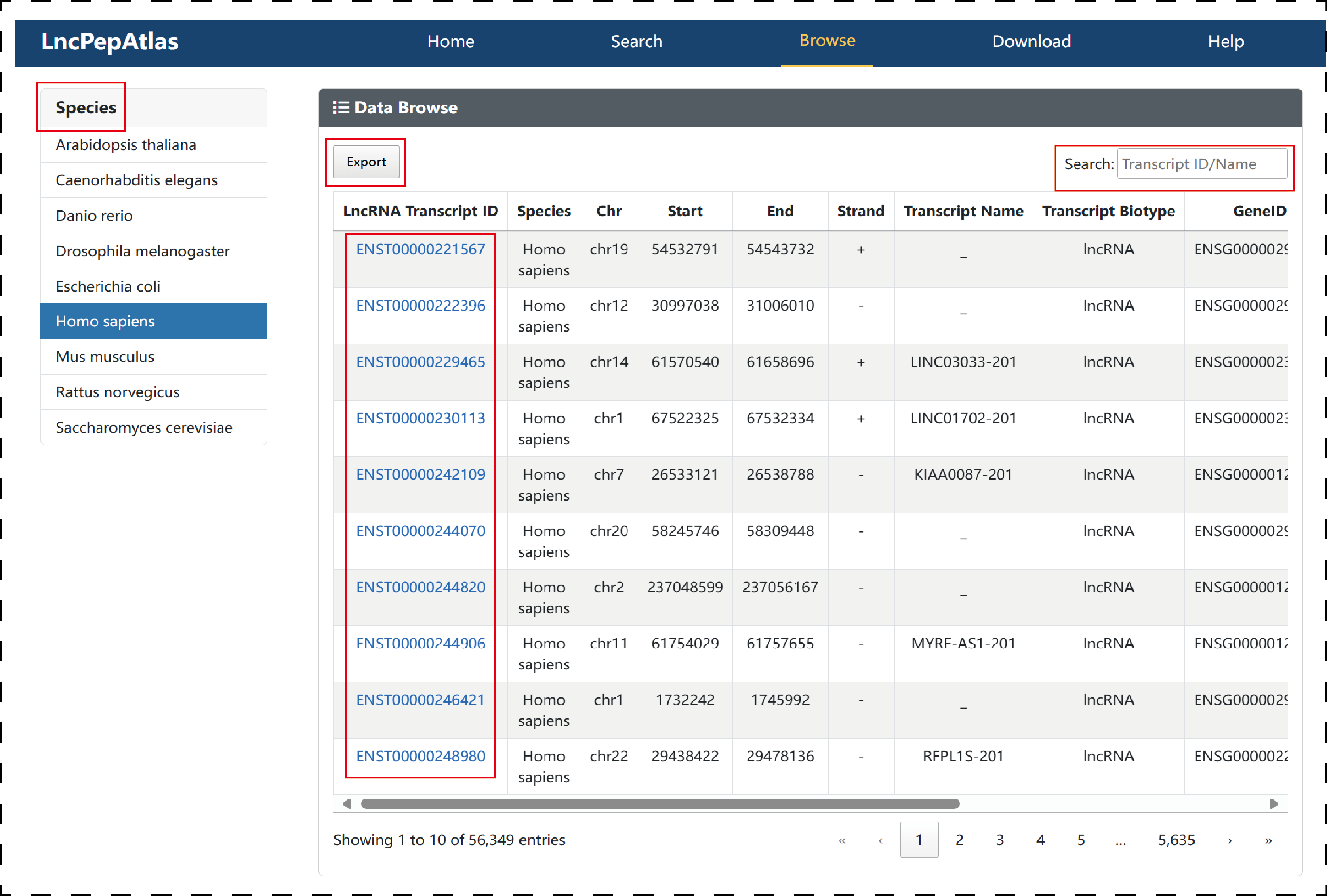
The ‘Browse’ page is organized as an interactive table that allows users to quickly search for the transcript of lncRNAs and customize filters according to ‘Species’. Users can click the ‘Export’ to download the basic information of lncRNAs and enter key information in ‘Search’ for fuzzy matching . To view details of a lncRNAs, users click on ‘LncRNA Transcript ID’.
4.2 Search
LncPepAtlas provides multi-type query modes to facilitate the users to query translatable lncRNAs and their corresponding peptides flexibly. The users can determine the scope of translatable lncRNAs and their corresponding peptides through ‘Search by Transcript ID’, ‘Search by Gene’ and ‘Search by Peptide’.
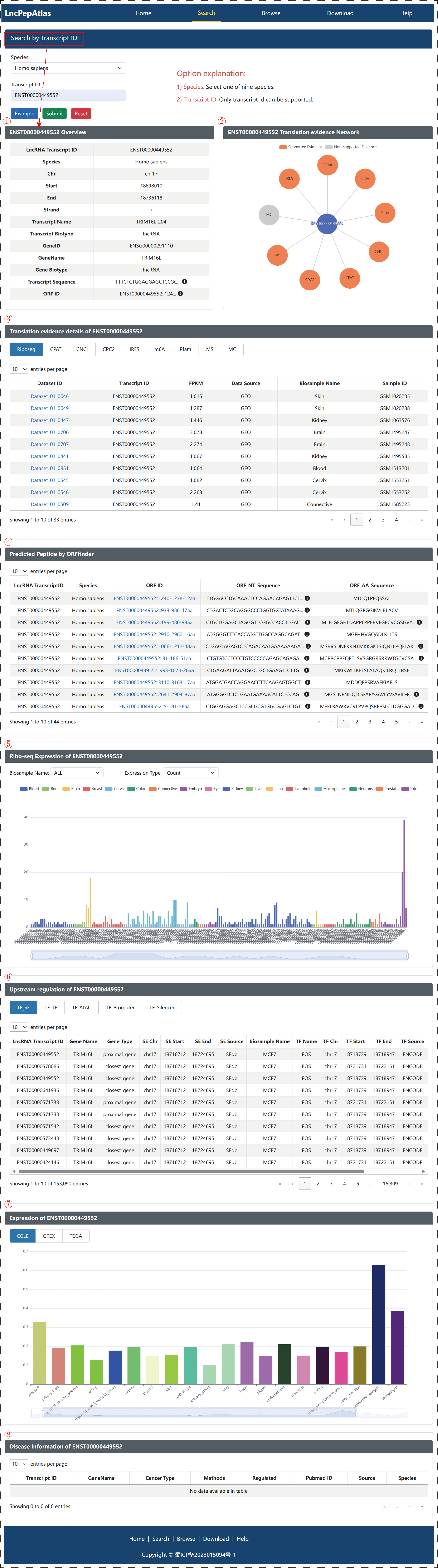
In ‘Search by Transcript ID’, the users can search translatable lncRNAs by selecting species and inputing transcript id. After submitting, the users can obtain the detailed information of on the input transcript id, including ‘LncRNA Overview’, ‘LncRNA Translation evidence Network’, ‘Translation evidence details of lncRNA’, ‘Predicted Peptide by ORFfinder’, ‘Ribo-seq Expression of LncRNA’, ‘Upstream regulation of LncRNA’, ‘Expression of LncRNA’ and ‘Disease Information of LncRNA’.
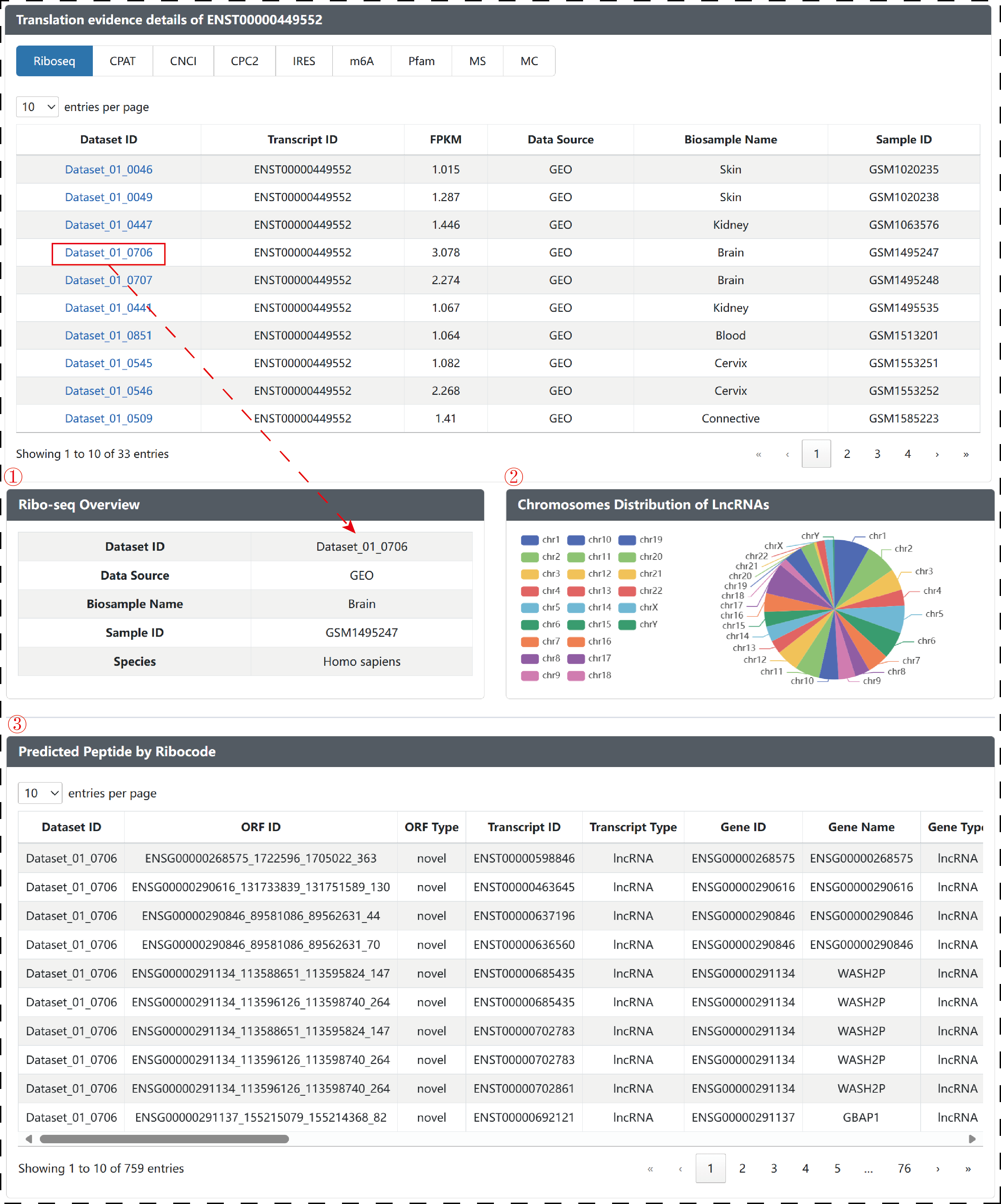
In ‘translation evidence details of transcript’ module, users can obtain ribosome profiling dataset detailed information, including ‘Ribo-seq Overview’, ‘Chromosomes Distribution of LncRNAs’ and ‘Predicted Peptide by Ribocode’
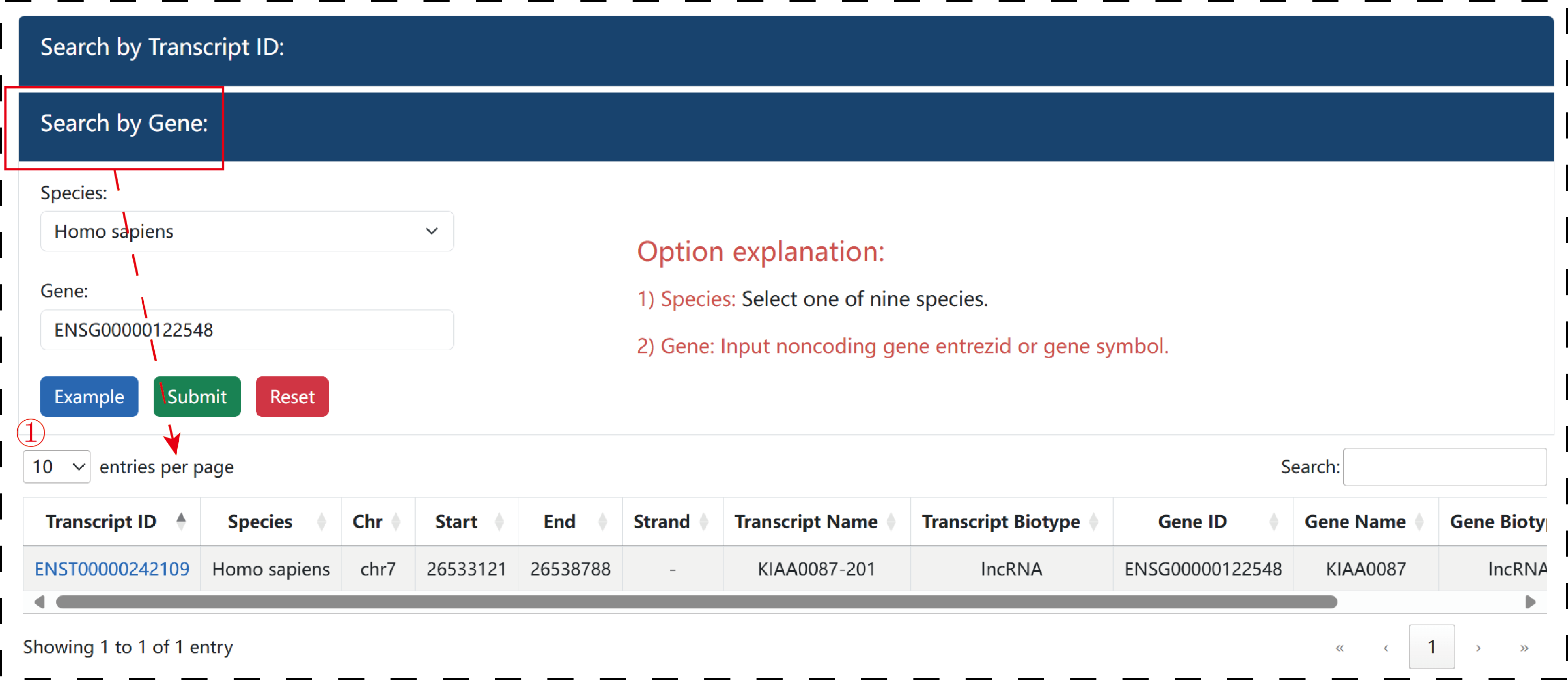
In ‘Search by Gene’, users could input an interesting gene entrez ID or gene symbol and select species, LncPepAtlas will return a result table of input gene related transcript information. Users hyperlink.
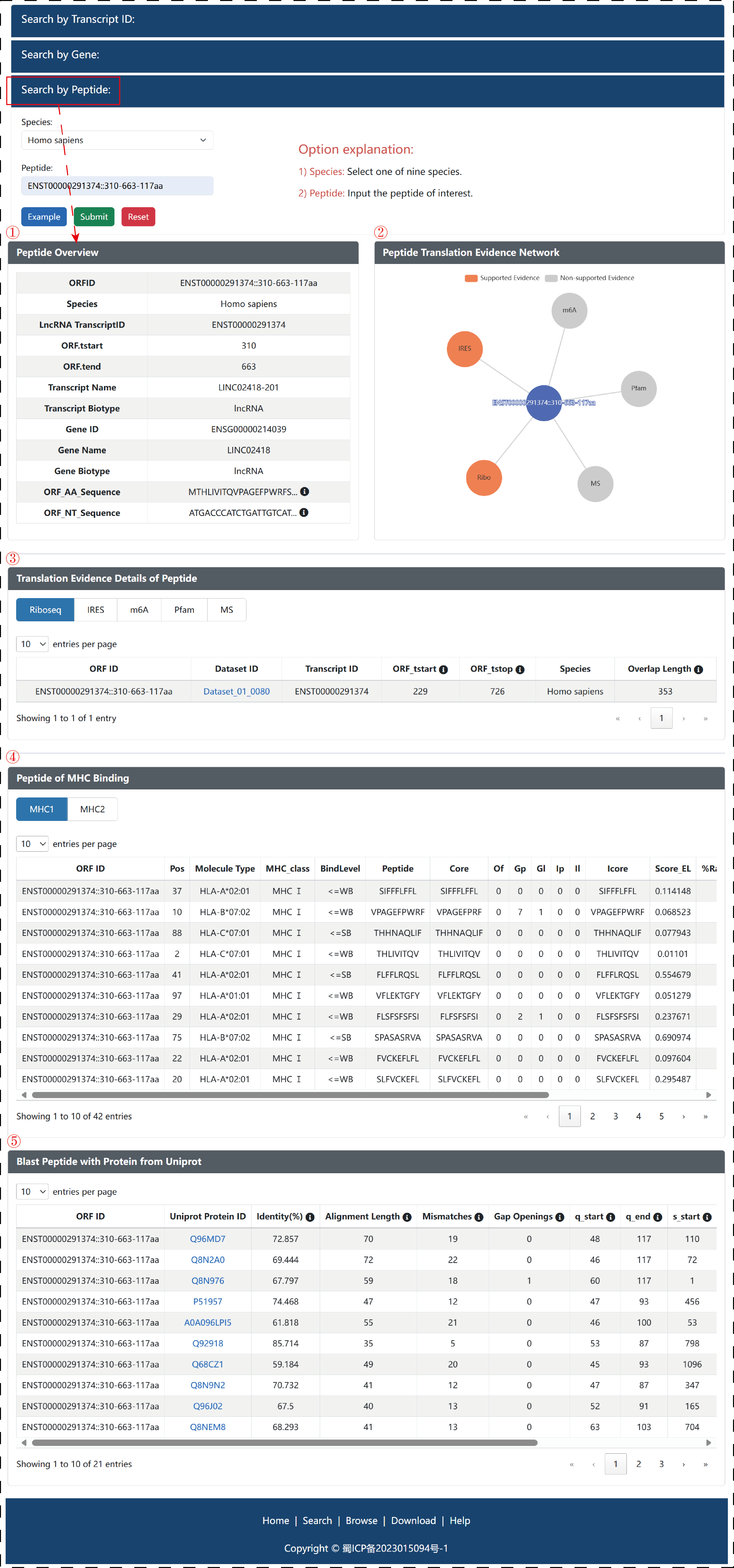
In ‘Search by Peptide’, Users can also select a kind of species and input an interesting peptide, and then LncPepAtlas will return five detailed module related input peptide, including ‘Peptide Overview’, ‘Peptide Translation Evidence Network’, ‘Translation Evidence Details of Peptide’, ‘Peptide of MHC Binding’ and ‘Blast Peptide with Protein from Uniprot’.
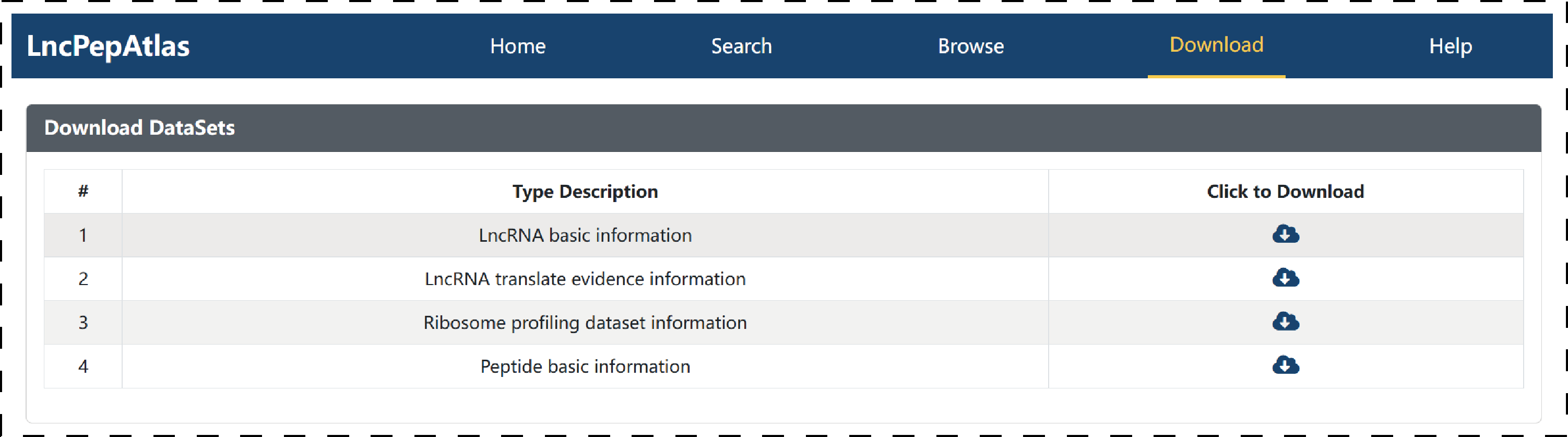
4.3 Download
LncPepAtlas provides a convenient and flexible download function for translatable lncRNAs-related files, including ‘LncRNA basic information’, ‘LncRNA translate evidence information’, and ‘Peptide basic information’.
The current version of LncPepAtlas backend operates on MySQL v5.7 and runs on Java Server Pages utilizing the Tomcat container v8.5.55. The frontend is a multi-page web application constructed with HTML, JavaScript, and CSS, incorporating the jQuery library v3.6.0 , Datatable v2.0.8 and ECharts v5.5.1 plugins. LncPepAtlas has been optimized for compatibility with major web browsers, including Google Chrome, Firefox and Apple Safari for the best display.